| The GOtcha method |
|
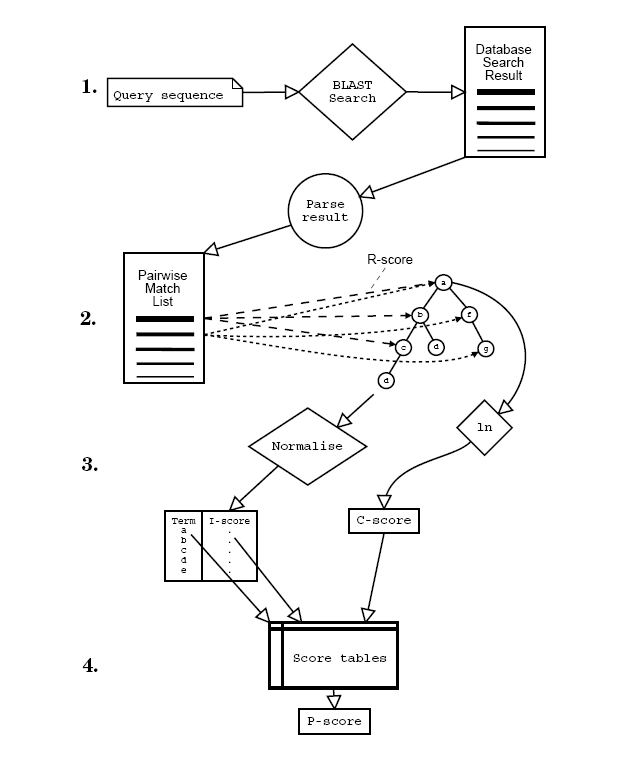
The GOtcha method was described in detail in Martin et al. BMC Bioinformatics (2004) 5:178. In summary, GOtcha assigns functional terms transitively based upon sequence similarity. The detailed method is outlined in the figure below.
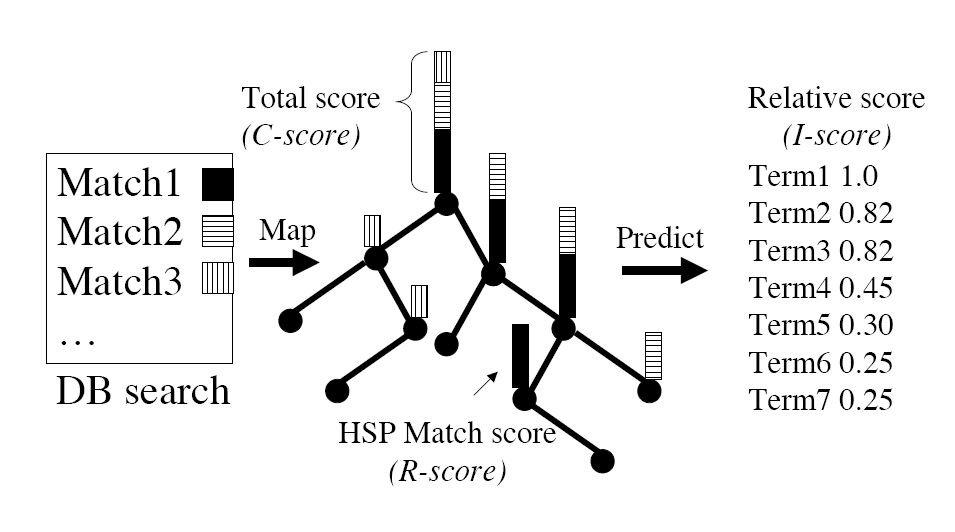
Scores on each node are assigned and accumulated as shown in the figure below.
Graphical output is provided using the Graphviz library, to provide a heat mapped graph of the assigned GO terms, colour coded by probability. |