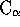 atoms taken
by averaging the coordinates for those positions within the file
ac_prot.8 that are found to be structurally equivalent. To obtain a poly
Alanine set of coordinates (i.e. including main chain and
atoms taken
by averaging the coordinates for those positions within the file
ac_prot.8 that are found to be structurally equivalent. To obtain a poly
Alanine set of coordinates (i.e. including main chain and 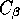 coordinates), type:
coordinates), type:




It may also be useful to have a set of averaged coordinates derived
from a protein structural family. This makes it possible to see what
portions of the structure are common to all members of the family (i.e.
the common core). The program AVESTRUC takes the output from STAMP
(i.e. an aligned family of protein structures), and generates a PDB file
containing averaged coordinates for the common core as identified by
STAMP. For example, to generate the averaged coordinates for the
aspartic proteinase domains one needs to type:
avestruc -f ac_prot.8 -o ac_prot_ave.pdb
The file ac_prot_ave.pdb will contain a set of averaged 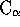 atoms taken
by averaging the coordinates for those positions within the file
ac_prot.8 that are found to be structurally equivalent. To obtain a poly
Alanine set of coordinates (i.e. including main chain and
atoms taken
by averaging the coordinates for those positions within the file
ac_prot.8 that are found to be structurally equivalent. To obtain a poly
Alanine set of coordinates (i.e. including main chain and 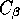 coordinates), type:
coordinates), type:
avestruc -f ac_prot.8 -o ac_prot_ave.pdb -polyA
Note that this will only work if all main chain atoms are found in the file
(i.e. it won't work if the PDB files contain only 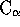 atoms).
atoms).