`-polyA' generate polyalanine model, the default is a C alpha model
`-c <STAMP char>' `-t <threshold>' `-w <window>'
these three parameters tell the program how to define structurally equivalent residues. `STAMP char' is the label of the STAMP field specified by the `#' character in the alignment file. `threshold' is the minimum (or maximum in the case of RMS deviation) value of the specified STAMP parameter tolerated, and `window' tells the minimum number of residues over which this must be true for structural equivalence. This is less complicated than it sounds.
The default is as described in [Russell & Barton, 1992]:
STAMP char = `G' (i.e.
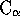 )
)threshold = 6.0
window = 3
(i.e. stretches of three or more residues having
 are considered equivalent)
are considered equivalent)`-alignment' this flag negates the effect of the previous three flags, and defines positions as equivalent if all structures are present at a position (i.e. positions not containing any gaps are deamed equivalent). Probably this is useful only for highly similar 3D structures.



