


Next: Using SCAN mode as
Up: Worked examples
Previous: Multiple alignment using an
Contents
Database scanning within STAMP is unpublished, apart from a brief description in a figure
legend [16], but it has been fairly well tested
since version 2.0. Indeed, two novel similarities have resulted
in publications [9,16].
Immunglobulin domain
One example of a scan is given. The light chain variable domain
of the immunoglobulin 2FB4 is used to scan a small database of
other protein domains containing both a diverse collection of
related folds (greek key folds, including azurin, superoxide
dismutase, CD4, etc.), and completely unrelated folds (such as
globins). See the directory examples/ig for this example.
The 2FB4 domain is described in 2fb4lv.domain. To scan this
against the database type:
stamp -l 2fb4lv.domain -s -n 2 -slide 5 -prefix 2fb4lv_stamp -d some.domains -cut
`-s' specifies the SCAN mode `-slide' describes how many residues to slide the
query sequence (2fb4lv) along each sequence in the file some.domains to provide
each initial fit (i.e. the sequence of 2fb4lv is layed on top of each database
sequence at postions 1, 6, 11, etc.). `-cut' tells the program to cut down each
domain read in from some.domains according to where the similarity is found.
If it is not specified, the output will contain domain descriptors identical to
those found in `some.domains'. When one is comparing a single-domain query to
a database structure having multiple domains, it is desirable to do this. Try running
it both ways (with and without -cut) and look at the output to see the difference.
(e.g. CHAIN A is converted to A 1 _ to A 60 _ in one descriptor in the SCAN
output and A 120 _ to A 175 _ in another, since there are two repeats of the
query domain in the database structure).
The above run should write the following to the
standard output (again, ignoring the header):
STAMP Structural Alignment of Multiple Proteins
Version 4.4 (May 2010)
by Robert B. Russell & Geoffrey J. Barton
Please cite PROTEINS, v14, 309-323, 1992
Results of scan will be written to file 2fb4lv_stamp.scan
Fits = no. of fits performed, Sc = STAMP score, RMS = RMS deviation
Align = alignment length, Nfit = residues fitted, Eq. = equivalent residues
Secs = no. equiv. secondary structures, %I = seq. identity, %S = sec. str. identity
P(m) = P value (p=1/10) calculated after Murzin (1993), JMB, 230, 689-694
(NC = P value not calculated - potential FP overflow)
Domain1 Domain2 Fits Sc RMS Len1 Len2 Align Fit Eq. Secs %I %S P(m)
Scan 2fb4lv 2fb4lc 1 4.317 2.120 111 105 127 55 46 8 10.87 78.26 1.00e+00
Scan 2fb4lv 2fb4l 1 9.799 0.001 111 166 111 111 111 11 100.00 97.30 0.00e+00
Scan 2fb4lv 1mcplv 1 7.848 1.165 111 113 116 96 95 0 49.47 40.00 2.05e-22
Scan 2fb4lv 1mcphv 1 6.921 1.500 111 122 126 85 81 0 30.86 34.57 1.44e-07
Scan 2fb4lv 1cmsC 1 2.507 1.639 111 148 157 28 24 4 4.17 62.50 1.00e+00
Scan 2fb4lv 3cd4 1 5.939 1.334 111 166 114 78 75 12 20.00 76.00 4.10e-03
Scan 2fb4lv 2hhbb 0 0.000 100.000 111 146 0 0 75 0 0.00 0.00 1.00e+00
Scan 2fb4lv 3dpa 0 0.000 100.000 111 166 0 0 75 0 0.00 0.00 1.00e+00
Scan 2fb4lv 3sgbe 0 1.940 2.313 111 166 204 25 17 3 5.88 88.24 1.00e+00
Scan 2fb4lv 1acx 1 4.152 2.454 111 108 133 57 43 4 16.28 72.09 7.26e-02
Scan 2fb4lv 2abxa 0 0.000 100.000 111 74 0 0 43 0 0.00 0.00 1.00e+00
Scan 2fb4lv 1l01 0 0.000 100.000 111 164 0 0 43 0 0.00 0.00 1.00e+00
Scan 2fb4lv 2azaa 1 4.063 2.463 111 129 134 49 35 5 14.29 82.86 1.00e+00
Scan 2fb4lv 1rnt 0 1.503 2.545 111 104 148 17 13 3 15.38 69.23 1.00e+00
Scan 2fb4lv 2sodo 1 3.611 2.365 111 151 158 42 32 8 9.38 71.88 1.00e+00
Scan 2fb4lv 2pcy 1 3.788 2.052 111 99 125 47 39 6 30.77 79.49 2.27e-04
Scan 2fb4lv 8atca 0 0.000 100.000 111 166 0 0 39 0 0.00 0.00 1.00e+00
See the file 2fb4lv_stamp.scan
where all of the fields are as for the PAIRWISE mode, save for Fits, which indicates the
number of fits that were saved to the file `2fb4lv_stamp.scan'. Note that for domain descriptors
(see some.domains) containing two Ig type folds (e.g. 2fb4l, 1cd4, etc.) that more than
one fit has been saved, since the search found both of the Ig type folds in each of
these two proteins. Not also that `Fits' is zero for several of the examples,
indicating that the no similarity was found within these proteins. Where more than one
Fit is output for a domain in the database, the best 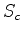 , RMS etc. are reported.
, RMS etc. are reported.
2fbjlv_stamp.scan will contain all the transformations output during
the scan. Several of these will be redundant, since it is possible for a
particular match to be found twice. To remove repeated
transformations, or those not considered interesting, run
the program SORTTRANS on the output.
sorttrans -f 2fb4lv_stamp.scan -s Sc 2.0 > 2fb4lv_stamp.sorted
This sorts the input file by 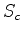 values, and leaves only those non-redundent
domain descriptions having an
values, and leaves only those non-redundent
domain descriptions having an
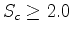 . A cutoff of
. A cutoff of 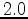 is generally
a good choice pairwise comparisons with a score lower than this tend to produce
poor quality alignments.
is generally
a good choice pairwise comparisons with a score lower than this tend to produce
poor quality alignments.
sorttrans -f 2fb4lv_stamp.scan -s rms 1.5 > 2fb4lv_stamp.sorted
sorts the input file by RMSD values, and leaves only those domain
descriptions having an RMSD 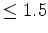 Å. Despite its predominance in
the literature, RMSD is not a very good means of measuring structural
similarity, since low RMSDs can usually be obtained for any two structures
if one considers a small enough set of residues.
Å. Despite its predominance in
the literature, RMSD is not a very good means of measuring structural
similarity, since low RMSDs can usually be obtained for any two structures
if one considers a small enough set of residues.
sorttrans -f 2fb4lv_stamp.scan -s nfit 40 > 2fb4l_stamp.sorted
sorts the input file by the number of atoms used in the final
fitting, and leaves only those domain descriptions where nfit 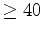 .
.
sorttrans -f 2fb4lv.scan -s n_sec 6 > 2fb4lv_stamp.sorted
sorts the input file by the number of equivalent secondary
structures, and leaves only those having 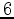 or more secondary
structures equivalent.
or more secondary
structures equivalent.
Combinations of these can be used to select out interesting domains
from a scan output. Probably the best combination involves Sc and
nfit (ie. score and nfit), since large structures can give
fortuitously large 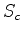 values with very few fitted atoms.
values with very few fitted atoms.
The final output is in the file 2fb4lv_stamp.sorted. This is
the result of the first example (i.e. -s Sc 2.0).
Note that several structures similar to the Ig type domain have
been detected, and appear (according to 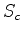 ) in the order one
might expect from knowledge of the 3D structures, sequences and
functions of these proteins.
) in the order one
might expect from knowledge of the 3D structures, sequences and
functions of these proteins.
The output from scanning can be used as input for other modes of
the program Once you have performed a scan, and have sorted the
`hits' down to an interesting set, you can then use the output from
scan as the input for a multiple alignment. This is discussed in the next section.



Next: Using SCAN mode as
Up: Worked examples
Previous: Multiple alignment using an
Contents