


Next: Transformations
Up: Input and Output format
Previous: Input and Output format
Contents
Every entry in a STAMP input file is called a `domain'. This term
is a bit of a misnomer, since `domains' needn't be single domains
(though it is usually best to do structure comparisons at the domain level).
The problem of defining domains such that a wide variety of
possibilities may be used (e.g. all the coordinates in a PDB file,
one chain, bits of one chain, two chains, one chain and bits of
another, etc) is solved by defining a domain by: 1) a file, 2) an
identifier, and 3) a list of `objects', from the file, to be
included in the domain. An object is defined as a run of
 coordinates, and a domain may contain more than one object.
coordinates, and a domain may contain more than one object.
Domains are stored in STAMP in files which may contain one or
more of such domain definitions.
The format of these files must be as follows:
<file name> <identifier label> { <objects> }
or,
<file name> <identifier label> { <objects> [RETURN]
R11 R12 R13 V1
R21 R22 R23 V2
R31 R32 R33 V3 }
 file name
file name is the full name (including path) of the PDB file in
which the coordinate information is to be found. If you don't know the
precise location of the file, then just call it UNK or something (i.e.
not a blank), and the programs should be able to find the appropriate PDB
file using the domain identifier field. Note that finding the PDB file
using the identifier relies on a set of rules defined in the
pdb.directories configuration file (see Chapter 5 for details of
the location of the file, its format and how STAMP uses it to locate
PDB files).
is the full name (including path) of the PDB file in
which the coordinate information is to be found. If you don't know the
precise location of the file, then just call it UNK or something (i.e.
not a blank), and the programs should be able to find the appropriate PDB
file using the domain identifier field. Note that finding the PDB file
using the identifier relies on a set of rules defined in the
pdb.directories configuration file (see Chapter 5 for details of
the location of the file, its format and how STAMP uses it to locate
PDB files).
 identifier label
identifier label is a short name to be used by the program.
eg. 4mbn1. The domain identifiers in a STAMP input file must be unique.
is a short name to be used by the program.
eg. 4mbn1. The domain identifiers in a STAMP input file must be unique.
DSSP secondary structure files can only be found by STAMP by using
the domain identifier in a similar fashion as described for PDB files
above. Again, see Chapter 5 for details of how this works.
 objects
objects are coordinate descriptions, and may be one of three types:
are coordinate descriptions, and may be one of three types:
1. ALL
all
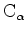 's from the file.
's from the file.
2. CHAIN X
only
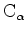 's labeled as chain X.
's labeled as chain X.
3. <chain1> <number1> <insert1> to <chain2> <number2> <insert2>
e.g. B 20 _ to B 67 P
only
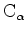 's between (and including) the two full brookhaven
's between (and including) the two full brookhaven
residue names (chain, number, insertion code;
the `_` character denotes a space)
N.B. THERE MUST BE AT LEAST ONE SPACE BETWEEN THE VARIOUS FIELDS.
Combinations of these are allowed within one domain, e.g. ` CHAIN A B 1
_ to B 65 _ `
R11  R33 and V1
R33 and V1  V3 are a rotation matrix and translation vector, respectively.
V3 are a rotation matrix and translation vector, respectively.
Thus, a full description of three domains might look something
like this:
/data/newpdb/pdb/pdb1ton.ent 1ton { ALL
0.9876 0.34 0.543 19.23
1.0 2.34 0.98473332 1.0
0.023 0.94 4.345 20.0 }
/data/newpdb/pdb/pdb2kai.ent 2kai_Kallikrien { CHAIN X CHAIN Y }
/data/newpdb/pdb/pdb3sgb.ent 3sgbe_SGprotease { E 20 _ to E 160 P
1.0 0.0 0.0 0.0
0.0 1.0 0.0 0.0
0.0 0.0 1.0 0.0 }
Note the spaces. There must be spaces separating each keyword or
datum to be read, even between the braces. For example:
/data/newpdb/pdb/pdb3sgb.ent 3sgb_protease{E 20 _ to E 160P}
would not be allowed.
In the second domain (Kallikrein) the transformation will be set equal
to the identity matrix with a translation of zero, since none has been
supplied.
The domains must be listed at the start of a file (ie. nothing must
come before them in a file), but anything may come afterwards,
provided that it contains no braces (ie. { or }) unless they are on lines
containing `%` in the first column.
It is possible to reverse the direction of an object in a domain
description. For example, if one has two objects,
one can reverse the direction of one or more of these by placing the
word "REVERSE" in front of the object, e.g.:
/data/newpdb/pdb/pdb4mbn.ent { REVERSE _ 1 _ to _ 20 _ _ 21 _ to _ 120 _ }



Next: Transformations
Up: Input and Output format
Previous: Input and Output format
Contents