



Next: Discussion
Up: Results
Previous: Comparison of Domain
The structures that the algorithm identified as correctly split can be
divided on the basis of the number of domains they contain. Table 4
summarises the number of occurrences of an n domain protein.
Single domain proteins are the largest group at 75% of the set.
Over the entire set there is an average of 1.3 domains
per protein. The number of occurrences of an n domain protein
falls off rapidly as n is increased and 98% of the proteins
contain three or fewer domains.
Examples of a single, two and four domain proteins are shown in
Figure 3.
Figure 3a shows
trypsin [Read & James, 1988], a serine protease. It is divided into two
domains, with a single cut in the middle of the chain and with both
the N and C termini crossing back over into opposite domains, making
each domain a two segment domain, similar to the topology of the two
domains shown in Figure 1c. Figure 3b illustrates the A chain of the
protein phosphoglucomutase [Lin et al., 1986]. It is split into four
domains. The chain runs from the first domain into the first half of
the second domain, passes through the third domain, comes back into the
second domain to complete it and finally makes up the fourth domain.
Figure 4 shows the distribution of the number of residues in a domain.
Most domains are made up of between 50
and 100 residues. 90% of the domains are comprised of less than
200 residues. The histogram tails off rapidly for large domains and
there are only two domains made up of more than 400 residues
(the two domains of glycogen phosphorylase).
Although the algorithm is primarily designed to search for single
segment or double segment domains, it is possible for domains to be
made up of more segments by non-contiguous `chopped bits' being added
onto the domain. Table 5 summarises the number of n segment
domains. 81.5% of the domains found were single segment. A further
17.6% of the domains were made up of two segments. Only one three
segment and one four segment domain were found in the final set (both
the domains of glucose oxidase [Hecht et al., 1993]).
The two segment domains were subclassified on the basis of those in
which there is a large difference in the relative sizes of the
segments. The size of the smaller segment as a percentage of the size
of the whole domain was calculated (histogram on diskette). The
distribution is fairly even over the entire range, though the
number of domains, in which one segment is 20-40% the size
of the other, is significant.
The distance separating the residue at the end of the first segment
and the residue at the start of the second segment was examined as a
percentage of the size of the intervening segment. The size of the
intervening segment was estimated by working out the maximum
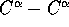 separation in the domain (histogram on diskette).
The distribution appears to be normal
with a peak in the range 30-40%. For 76% of the domains the
separation is less than half the maximum
separation in the domain (histogram on diskette).
The distribution appears to be normal
with a peak in the range 30-40%. For 76% of the domains the
separation is less than half the maximum 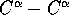 separation in the intervening segment. This shows that most inserted
domains have their connections to the rest of the protein close
together. A close connection may suggest that the inserted domain
could be deleted without disrupting the integrity of the two segment
domain.
separation in the intervening segment. This shows that most inserted
domains have their connections to the rest of the protein close
together. A close connection may suggest that the inserted domain
could be deleted without disrupting the integrity of the two segment
domain.
No correlation was found between the end point distance and the
relative sizes of the segments.




Next: Discussion
Up: Results
Previous: Comparison of Domain
as@bioch.ox.ac.uk
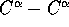 separation in the domain (histogram on diskette).
The distribution appears to be normal
with a peak in the range 30-40%. For 76% of the domains the
separation is less than half the maximum
separation in the domain (histogram on diskette).
The distribution appears to be normal
with a peak in the range 30-40%. For 76% of the domains the
separation is less than half the maximum 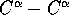 separation in the intervening segment. This shows that most inserted
domains have their connections to the rest of the protein close
together. A close connection may suggest that the inserted domain
could be deleted without disrupting the integrity of the two segment
domain.
separation in the intervening segment. This shows that most inserted
domains have their connections to the rest of the protein close
together. A close connection may suggest that the inserted domain
could be deleted without disrupting the integrity of the two segment
domain.



