



Next: Alignment using an
Up: Worked examples
Previous: Worked examples
Mammalian and Bacterial Serine Proteinases
(This example is discussed in Russell & Barton, (1992).)
Despite a pronounced functional similarity (a highly conserved catalytic
triad), this family of proteins shows little overall sequence similarity.
Indeed, sequence alignment methods generally fail to provide an accurate
alignment of these protein sequences. In situations like these, STAMP
can be used to provide an accuarate alignment of protein sequences based
on a comparison of 3D structure. This can often reveal regions of weak
sequence similarity that are not detectable during a comparison of
sequence. The files for this example are in the directory examples/s_prot in the
directory where you have installed STAMP.
A list of the domains is given in the file s_prot.domains. The
output. Running PDBSEQ:
pdbseq -f s_prot.domains > s_prot.seqs
produced the file s_prot.seqs, from which an AMPS multiple
sequence alignment was produced, and stored in the file
s_prot_amps.align. Running ALIGNFIT:
alignfit -f s_prot_amps.align -d s_prot.domains -out s_prot_alignfit.trans
should give the output:
ALIGNFIT R.B. Russell 1995
Reading in block file...
Blocfile read: Length: 261
Reading in coordinate descriptions...
Reading coordinates...
Checking for inconsistencies...
Doing pairwise comparisons...
Doing treewise comparisons...
ALIGNFIT done.
Look in the file s_prot_alignfit.trans for output and details
The final transformation
(called alignfit.trans if default ALIGNFIT settings are used) is
in the file s_prot_alignfit.trans.
This provides an initial set of transformations for use by
STAMP. To run STAMP type:
stamp -l s_prot_alignfit.trans -prefix s_prot
Should produce the following output:
STAMP Structural Alignment of Multiple Proteins
by Robert B. Russell & Geoffrey J. Barton
Please cite PROTEINS, v14, 309-323, 1992
Sc = STAMP score, RMS = RMS deviation, Align = alignment length
Len1, Len2 = length of domain, Nfit = residues fitted
Secs = no. equivalent sec. strucs. Eq = no. equivalent residues
%I = seq. identity, %S = sec. str. identity
No. Domain1 Domain2 Sc RMS Len1 Len2 Align NFit Eq. Secs. %I %S
Pair 1 4chaa 3est 7.68 1.12 239 240 241 207 205 20 36.67 79.58
Pair 2 4chaa 2ptn 7.74 1.01 239 223 233 200 199 20 41.42 82.01
Pair 3 4chaa 1ton 6.82 1.28 239 227 241 182 178 19 31.80 76.57
Pair 4 4chaa 3rp2a 7.40 1.15 239 224 234 194 187 18 30.96 75.31
Pair 5 4chaa 2pkaab 7.21 1.34 239 232 240 197 194 19 31.38 80.75
Pair 6 4chaa 1sgt 7.04 1.33 239 223 238 189 182 20 30.13 79.92
<etc.>
Reading in matrix file s_prot.mat...
Doing cluster analysis...
Cluster: 1 ( 2ptn & 2pkaab ) Sc 8.50 RMS 1.08 Len 232 nfit 213
See file s_prot.1 for the alignment and transformations
Cluster: 2 ( 2sga & 3sgbe ) Sc 8.36 RMS 0.62 Len 191 nfit 164
See file s_prot.2 for the alignment and transformations
<etc.>
Cluster: 8 ( 1sgt & 4chaa 3est 3rp2a 1ton 2ptn 2pkaab )
Sc 7.58 RMS 1.14 Len 267 nfit 177
See file s_prot.8 for the alignment and transformations
Cluster: 9 ( 1sgt 4chaa 3est 3rp2a 1ton 2ptn 2pkaab
& 2alp 2sga 3sgbe ) Sc 4.79 RMS 1.86 Len 292 nfit 109
See file s_prot.9 for the alignment and transformations
The various fields describe details of the pairwise and treewise
comparisons: 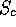 , RMS deviation, the alignment length (Align),
the length of each structure in residues (Len1, Len2), the number of
atoms used in the RMS fit (Nfit), the number of equivalent secondary
structure elements (Secs), and the number of equivalent residues
(see above, Eq.).
, RMS deviation, the alignment length (Align),
the length of each structure in residues (Len1, Len2), the number of
atoms used in the RMS fit (Nfit), the number of equivalent secondary
structure elements (Secs), and the number of equivalent residues
(see above, Eq.).
STAMP will also produce several files:
s_prot.mat -- a file containing the information used to derive the
structural similarity tree (i.e. the output from the PAIRWISE) mode. This
is an upper diagonal matrix containing the pairwise 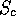 values.
values.
s_prot.N -- a series of files containing transformations and alignments
created by running the TREEWISE mode in STAMP. Each file corresponds to
a node in the similarity tree (i.e. a cluster), where two
groups of one or more structures have been combined to form an
alignment and transformations. The higher the value of N the more
structurally dissimilar the proteins contained in the file are. Highly
similar structures are clustered (aligned/superimposed) at an early stage
in the program, with more distantly related structures being clustered
towards the end.
The top of each file contains the information needed to generate (using
TRANSFORM, see below)
superimposed coordinates (in STAMP domain format, see below).
After these details, various details of the similarity (RMS deviation,
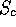 value, etc) are given. The bottom portion of the file contains
the structural alignment in STAMP format. Positions not aligned with
gaps contain information as to the degree of local structural similarity,
such as the distance between (averaged)
value, etc) are given. The bottom portion of the file contains
the structural alignment in STAMP format. Positions not aligned with
gaps contain information as to the degree of local structural similarity,
such as the distance between (averaged) 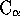 atoms, and the
atoms, and the
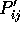 value.
value.
Methods for displaying sequence alignments and structures are described below.




Next: Alignment using an
Up: Worked examples
Previous: Worked examples
Rob Russell and Geoff Barton
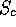 , RMS deviation, the alignment length (Align),
the length of each structure in residues (Len1, Len2), the number of
atoms used in the RMS fit (Nfit), the number of equivalent secondary
structure elements (Secs), and the number of equivalent residues
(see above, Eq.).
, RMS deviation, the alignment length (Align),
the length of each structure in residues (Len1, Len2), the number of
atoms used in the RMS fit (Nfit), the number of equivalent secondary
structure elements (Secs), and the number of equivalent residues
(see above, Eq.).



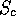 values.
values.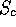 value, etc) are given. The bottom portion of the file contains
the structural alignment in STAMP format. Positions not aligned with
gaps contain information as to the degree of local structural similarity,
such as the distance between (averaged)
value, etc) are given. The bottom portion of the file contains
the structural alignment in STAMP format. Positions not aligned with
gaps contain information as to the degree of local structural similarity,
such as the distance between (averaged) 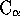 atoms, and the
atoms, and the
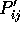 value.
value.