



Next: Database Scanning
Up: Worked examples
Previous: Multiple alignment (PAIRWISE
This method avoids having to create an initial sequence alignment, and
tends to work for homologous proteins, or those having very similar
lengths despite no sequence similarity.
Globins
Since the globin sequences are of similar length an initial
superimposition accurate enough to proceed with STAMP can be
obtained by merely aligning the N-terminal ends of the sequences
and using whatever equivalences result to obtain an initial
superimposition. The command ROUGH (ROUGHFIT procedure) is used. In addition,
an initial conformation based fit is performed
in order that any inaccuracies in this initial superimposition may be corrected.
See the directory examples/globins.
To run STAMP in this example, type:
stamp -l globin.domains -rough -n 2 -prefix globin
should produce the following on the standard output (ignoring the header):
Running roughfit.
Sc = STAMP score, RMS = RMS deviation, Align = alignment length
Len1, Len2 = length of domain, Nfit = residues fitted
Secs = no. equivalent sec. strucs. Eq = no. equivalent residues
%I = seq. identity, %S = sec. str. identity
No. Domain1 Domain2 Sc RMS Len1 Len2 Align NFit Eq. Secs. %I %S
Pair 1 2hhbb 2hhba 8.08 1.37 146 141 146 133 130 7 41.10 78.08
Pair 2 2hhbb 2lhb 7.01 1.49 146 149 150 125 124 7 24.16 77.18
Pair 3 2hhbb 4mbn 7.96 1.42 146 153 147 138 137 8 23.53 77.78
Pair 4 2hhbb 1ecd 6.79 2.10 146 136 143 122 114 7 17.12 76.71
Pair 5 2hhbb 1lh1 5.80 2.39 146 153 154 112 106 7 15.69 69.28
<etc.>
Cluster: 5 ( 1lh1 & 2lhb 1ecd 4mbn 2hhbb 2hhba )
Sc 7.83 RMS 2.45 Len 156 nfit 116
See file globin.5 for the alignment and transformations
where the output and files are as described for the serine proteinase example above,
with `s_prot' replaced with `globin'.
-rough performs the initial superimpositions (ROUGHFIT) and -n 2 means that the conformation
biased fit will be performed before the final fit. This conformation biased fit is
usually necessary when the initial superimpositions are approximate.
ROUGHFIT will not always work. Note that in this example all the pairwise
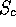 values are above
values are above 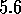 , suggesting strong structural similarity. If
when using the ROUGHFIT option you find low
, suggesting strong structural similarity. If
when using the ROUGHFIT option you find low 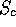 values (the program will
cry out LOW SCORE -- see the manual), this usually means that ROUGHFIT hasn't
managed to generate a good enough starting superimposition, and you should
try something else, such as is described in the next section.
values (the program will
cry out LOW SCORE -- see the manual), this usually means that ROUGHFIT hasn't
managed to generate a good enough starting superimposition, and you should
try something else, such as is described in the next section.
Rob Russell and Geoff Barton
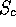 values are above
values are above 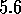 , suggesting strong structural similarity. If
when using the ROUGHFIT option you find low
, suggesting strong structural similarity. If
when using the ROUGHFIT option you find low 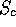 values (the program will
cry out LOW SCORE -- see the manual), this usually means that ROUGHFIT hasn't
managed to generate a good enough starting superimposition, and you should
try something else, such as is described in the next section.
values (the program will
cry out LOW SCORE -- see the manual), this usually means that ROUGHFIT hasn't
managed to generate a good enough starting superimposition, and you should
try something else, such as is described in the next section.



