



Next: Allowing for Two
Up: Materials and Methods
Previous: Introduction - Split
Consider chopping the protein chain
into two parts of segments between residues i and 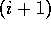 . A
segment can consist of any number of residues, but the residues must
form a continuous sequence along the chain. Segment A then
consists of residues 1 to i and segment B
of residues
. A
segment can consist of any number of residues, but the residues must
form a continuous sequence along the chain. Segment A then
consists of residues 1 to i and segment B
of residues 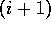 to N, where N is the number of residues
in the chain. The split value can then be calculated for
to N, where N is the number of residues
in the chain. The split value can then be calculated for 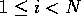 . Figure 5 illustrates a graph of split value against i for the
T-cell surface glycoprotein, CD4 [Ryu et al., 1990]. The split value has a
large peak at i = 97, indicating that the protein should be split
into two domains at this point. Once split, the two domains can
themselves be individually scanned to find the maximum split values
and hence the best positions to split them into new domains, which
again can be scanned and split and so on. By placing a limit on the
minimum number of residues in a domain (minimum domain size, MDS)
and/or defining a minimum split value (MSV) below which
the two parts are considered
to be correlated and not divisible into smaller domains,
the process of division can be stopped. The result is a series of
`cuts' defining how the chain should be split into separate domains.
. Figure 5 illustrates a graph of split value against i for the
T-cell surface glycoprotein, CD4 [Ryu et al., 1990]. The split value has a
large peak at i = 97, indicating that the protein should be split
into two domains at this point. Once split, the two domains can
themselves be individually scanned to find the maximum split values
and hence the best positions to split them into new domains, which
again can be scanned and split and so on. By placing a limit on the
minimum number of residues in a domain (minimum domain size, MDS)
and/or defining a minimum split value (MSV) below which
the two parts are considered
to be correlated and not divisible into smaller domains,
the process of division can be stopped. The result is a series of
`cuts' defining how the chain should be split into separate domains.
as@bioch.ox.ac.uk
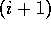 . A
segment can consist of any number of residues, but the residues must
form a continuous sequence along the chain. Segment A then
consists of residues 1 to i and segment B
of residues
. A
segment can consist of any number of residues, but the residues must
form a continuous sequence along the chain. Segment A then
consists of residues 1 to i and segment B
of residues 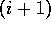 to N, where N is the number of residues
in the chain. The split value can then be calculated for
to N, where N is the number of residues
in the chain. The split value can then be calculated for 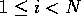 . Figure 5 illustrates a graph of split value against i for the
T-cell surface glycoprotein, CD4 [Ryu et al., 1990]. The split value has a
large peak at i = 97, indicating that the protein should be split
into two domains at this point. Once split, the two domains can
themselves be individually scanned to find the maximum split values
and hence the best positions to split them into new domains, which
again can be scanned and split and so on. By placing a limit on the
minimum number of residues in a domain (minimum domain size, MDS)
and/or defining a minimum split value (MSV) below which
the two parts are considered
to be correlated and not divisible into smaller domains,
the process of division can be stopped. The result is a series of
`cuts' defining how the chain should be split into separate domains.
. Figure 5 illustrates a graph of split value against i for the
T-cell surface glycoprotein, CD4 [Ryu et al., 1990]. The split value has a
large peak at i = 97, indicating that the protein should be split
into two domains at this point. Once split, the two domains can
themselves be individually scanned to find the maximum split values
and hence the best positions to split them into new domains, which
again can be scanned and split and so on. By placing a limit on the
minimum number of residues in a domain (minimum domain size, MDS)
and/or defining a minimum split value (MSV) below which
the two parts are considered
to be correlated and not divisible into smaller domains,
the process of division can be stopped. The result is a series of
`cuts' defining how the chain should be split into separate domains.



