Bioinformatics services powered by the Slivka framework
Run production DRSASP pipelines on our hosted Slivka-Bio server and explore the same open framework for packaging your own bioinformatics tools as resilient web services.
Slivka-Bio
Access services for multiple sequence alignment, structure predictions, disorder analysis, and conservation scoring provided by the DRSASP.
Slivka Framework
Publish command-line tools as managed web services with REST APIs, monitoring, and scheduler integration.
Slivka-Bio @ University of Dundee • Part of DRSASP • Backend for Jalview 2.12 web services
Run bioinformatics jobs on Slivka-Bio
This public Slivka-Bio deployment, maintained by the Barton Group within the Division of Computational Biology, delivers production-scale web services for DRSASP users worldwide.
Submit jobs for multiple sequence alignment, structure prediction, disorder analysis, or conservation scoring — all backed by the University of Dundee HPC cluster.
Available tools
- Multiple sequence alignment: Clustal Omega, Clustal W, MAFFT, MUSCLE, T-Coffee, ProbCons, MSAProbs
- Secondary structure predictions via JPred
- Disorder prediction: GlobPlot, DisEMBL, IUPred, JRONN
- Conservation analysis with AACon
How to use
- Submit via web forms.
- Use the Python client, or try the Colab demo.
- Explore the REST API for programmatic access.
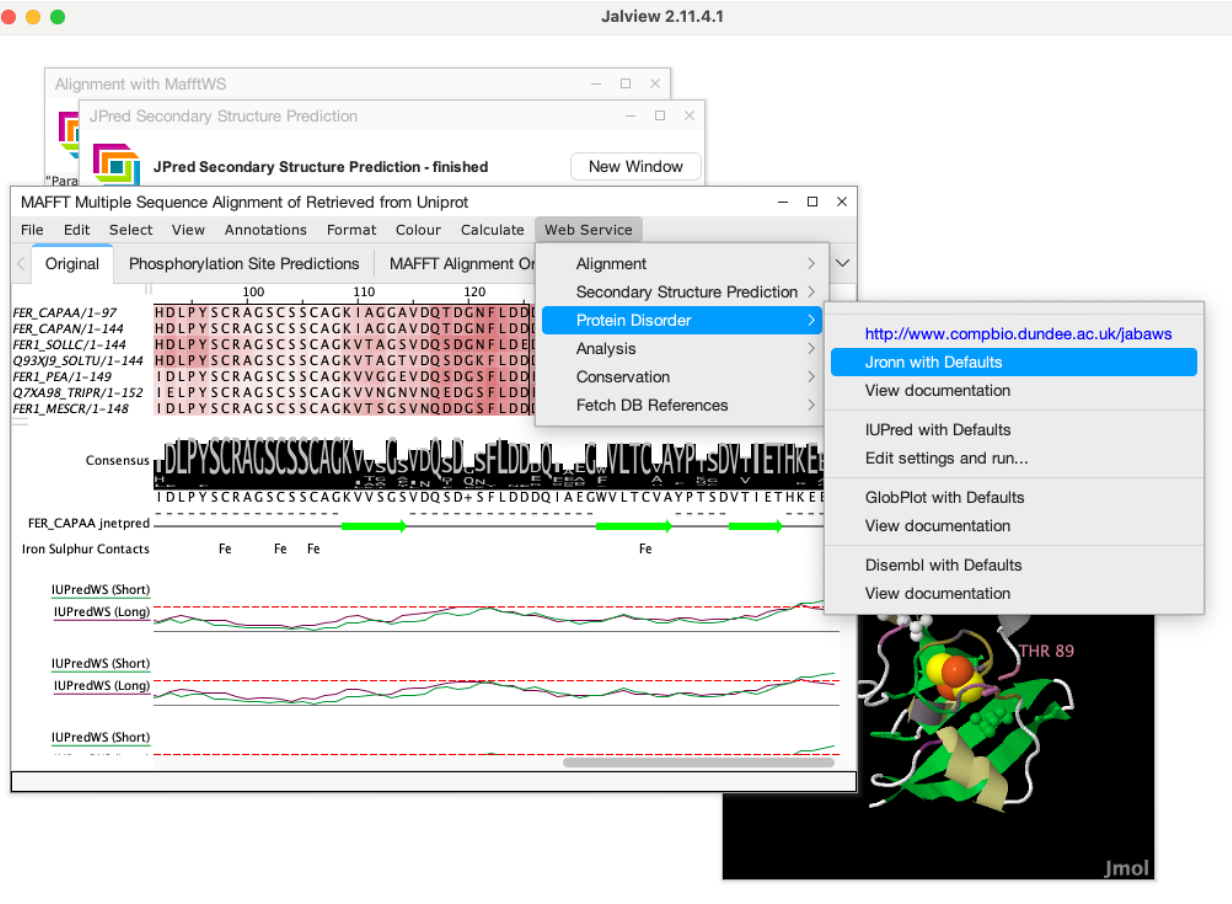
Jalview 2.12 connects directly to Slivka-Bio web services for alignments, structure prediction, and analysis.
Transform bioinformatics tools into scalable web services
Slivka turns command-line tools into scalable, user-friendly web services. Using straightforward YAML configurations, it creates RESTful APIs, oversees HPC job scheduling, and provides real-time job monitoring.
Deploy Slivka anywhere with the installer or Docker stack to unlock reproducible, production-ready bioinformatics services.
- CLI → REST API with one YAML file
- Supports local execution, HPC, and cloud services
- Slivka-Bio Docker image for containerised deployments
- Python framework and service installer for local clusters
The Slivka framework enables rapid deployment of new services, ensuring that researchers have access to the latest bioinformatics tools through consistent interfaces.
Integrated with Jalview and DRSASP
Slivka-Bio powers the web services in Jalview 2.12 and the Dundee Resource for Sequence Analysis and Structure Prediction (DRSASP).
Jalview 2.12 can automatically discover new Slivka services, allowing developers to deploy tools that researchers can immediately integrate with sequence, homology and structure data.
The DRSASP is transitioning to a Slivka-Bio backend to streamline service deployment. Check out the development Slivka-Bio instance to see our latest updates.
Slivka Architecture
Slivka's modular design separates the REST API from job scheduling and execution, enabling seamless integration with local runners, HPC schedulers, or cloud compute.
From YAML to live API
Define a service once and Slivka handles the rest: validation, data capture, job dispatch, monitoring, and results delivery. The example shown here is the same configuration powering our public services.
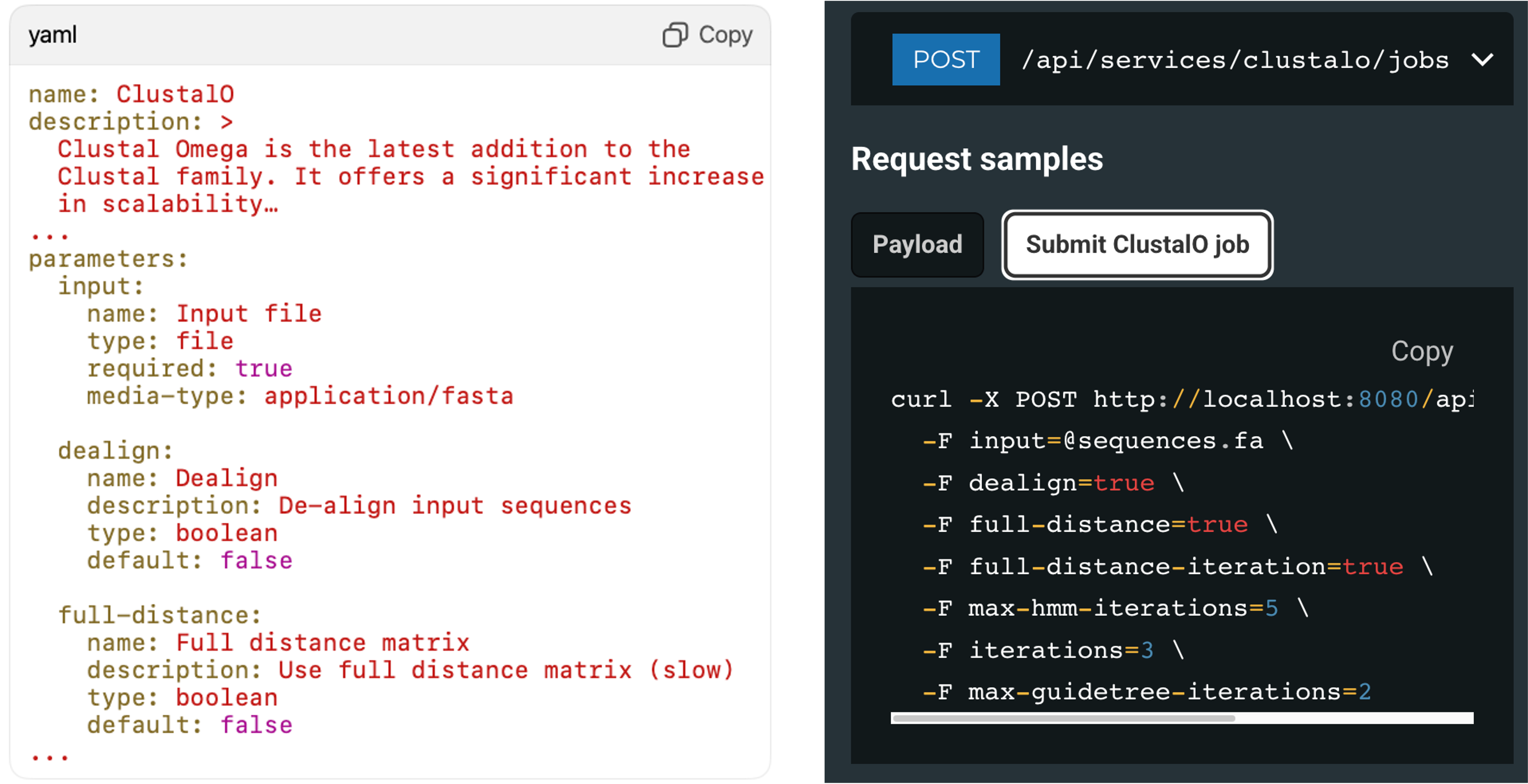
A simple YAML service description results in a functional API, executing jobs on your local compute or HPC scheduler. Configuration files define inputs, validation rules, and command templates.
Example service configs.Slivka Ecosystem
Docker Stack
Deploy a preconfigured Slivka-Bio instance for instant exploration or testing.
Docker Hub →Installer
Install Slivka and manage services on your infrastructure using the Python installer.
Installer Repo →Czekolada Client
Launch the Node.js client for a rich GUI that submits and tracks Slivka jobs.
Czekolada on GitHub →About Slivka-Bio
Slivka-Bio is developed and operated by the Barton Group in the Division of Computational Biology at the University of Dundee in collaboration with partners at Genentech. The platform is shaped by the Slivka framework, with key contributions from Mateusz Warowny, Thomas Down, Stuart MacGowan, Kiran Mukhyala, Geoff Barton, and James Procter.
Together with the Dundee Resource for Sequence Analysis and Structure Prediction (DRSASP), the service provides resilient, production-ready access to community bioinformatics tools and web services.
Contact & Citation
We welcome collaboration and feedback on Slivka-Bio and the Slivka framework. Reach the team at the University of Dundee via the Barton Group.
The Barton GroupDivision of Computational Biology
School of Life Sciences
University of Dundee
Dow Street
Dundee, DD1 5EH
Scotland, UK
Recommended Citation
Stuart A. MacGowan, Fábio Madeira, Thiago Britto-Borges, Mateusz M. Warowny, Alexey Drozdetskiy, James B. Procter, and Geoffrey J. Barton, The Dundee Resource for Sequence Analysis and Structure Prediction, Protein Science (2019). https://doi.org/10.1002/pro.3783
