


Next: ALIGNFIT
Up: Summary of parameters for
Previous: PDB checker (PDBC)
Contents
This program takes a list of protein domains (ie. a LISTFILE) and
outputs a series of sequences derived from the described PDB
files. The format is:
pdbseq -f <domain file> [-min <val> -max <val> -separate
-foramt <fasta> -v -tl <max title length>]
`-min/max 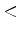 val
val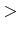 ' specify the minimum/maximum sequence length
to be output. If the length of a sequence is less than min or greater than max, the
sequence will be skipped (useful particularly if one wants to ignore very short
PDB sequence, such as peptide inhibitors, etc.).
' specify the minimum/maximum sequence length
to be output. If the length of a sequence is less than min or greater than max, the
sequence will be skipped (useful particularly if one wants to ignore very short
PDB sequence, such as peptide inhibitors, etc.).
The output is in NBRF (PIR) format, and is written to the standard
output. Using `-format  fasta
fasta will make the output as FASTA
format.
will make the output as FASTA
format.
The option `-separate' will produce files for each domain in the input file.
These files are named `ID'.seq.
The program outputs a title line that attempts to describe the protein
sequence according to the definitions given in the PDB file. The
TITLE, COMPND and SOURCE lines are strung together (in that order).
The option -tl <number> (tl = title limit) specifies the maximum length of this string.
This description will always be postfixed (after a ``:'') by
the range of residues considered (i.e. All, Chain a, etc.).



Next: ALIGNFIT
Up: Summary of parameters for
Previous: PDB checker (PDBC)
Contents