- Contacts
- Quick Search
- Protein Sequence Search
- BLAST Search
- Multiple Protein Search
- Table of Protein Interactions
- Evidence of Interaction
- Protein Summary
- Protein Network Viewer
|
Contact Details
If you have any problems or queries with this website, please send an email to . Thank you |
|
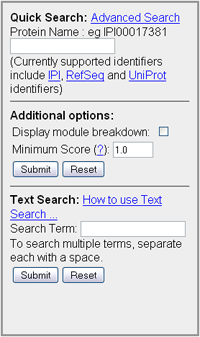 |
Quick Search
With Quick Search on the main page you can enter either an IPI (eg IPI00017381), RefSeq (eg NP_002907) or Uniprot (eg P35249) value to reference a protein within the database. Once you have clicked search you will be returned with the Protein Interactions results page. There are extra options that are also available to filter the results that are returned. These options include:
There is also a text search feature allowing searches based on the name or description of the protein to help locate the required protein. Query Syntax
It is also possible to indicate terms that you do not want to be included in the search results. Once you have listed the terms that you want to search for use the "NOT" command then type each term that is not to be included in the output. For example the text search query:
|
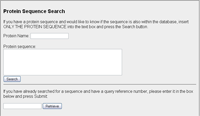 |
Protein Sequence Search
If you have a sequence for a protein, but don't have a valid accession, you can enter the sequence in the text box, giving it with a label and have it searched against the sequences that are in the database. Initially an exact sequence match is done. If this does not provide any results then you can submit the sequence to be aligned against all human protein sequences using BLASTp. From this you are given a unique reference key (e.g. P61417022). This unique key can be entered in the lower box to retrieve BLAST results from a previous request or one that you are returning to after waiting for the results. |
    |
BLAST Search
If your sequence does not match any that are currently in the database you are presented with a box to submit your sequence to a BLAST search. This should find any homologs of your protein that are in the database. Once your sequence has been submitted to BLAST you are presented with a page to retrieve your results. If you want to get them now, click on the Retrieve Results button. If not take a note of your reference tag, as this can be entered on the Search page to collect previously processed results results. If you have requested to retrieve your results they will be presented in a table listing your reference tag, the protein name, the percentage Identity, alignment length and e-Value as calculated by BLAST along with the number of predicted interactions greater than the selected Scores. The values in these columns link you through to those interactions. If your BLAST results are not ready for collection, the page will refresh every 10 seconds until the results are ready for collection. There is also the option to force a refresh of the page. |
  |
Multiple Protein Search
For a multiple protein search, insert the IPI tag for each protein separated by a single space into the box provided (image top left) on the search page. Then click Search. You will be returned with a results page that lists all of the sequences that you have entered (image botton left). If data relating to your sequence has been found then a link to the Protein Identifier page will be highlighted and the number of predicted interactors at pre-set Score cut-offs will be listed. These link through to a table showing the protein and the predicted proteins that it interacts with at the specified minimum interaction score. |
|
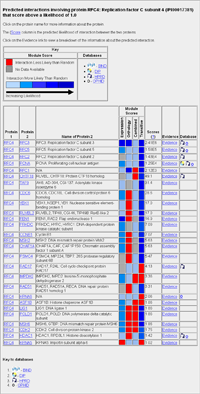
Table of Protein Interactions
Once your protein has been identified within the database, all proteins that have a predicted interaction with that protein are listed in a table shown top left. The normal results will just show your protein, the protein it is predicted to interact with and the final Score for that interaction. A flag is also displayed to identify if that interaction is present in external databases (BIND, DIP, HPRD or OPHID). If you selected "Displaying module breakdown" from the quick search box then you will get the indiviual scores for all of the predictive modules along with the total Score. Clicking on the protein identification name (e.g. RFC4) links to a page providing more details about that protein, including sequence, number of interactions with that protein and links to external databases. Clicking on the Evidence link generates a page that details the information that was used to calculate the Score value. |
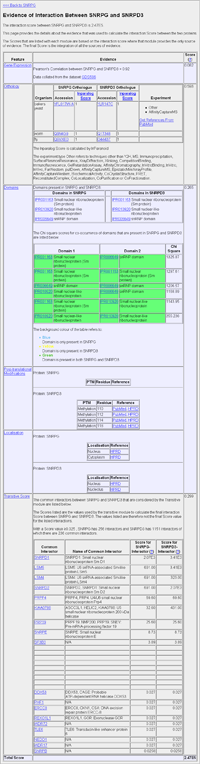 |
Evidence of Interaction
Due to the size of the page, the image (left) links to an example Evidence of Interaction webpage rather than an image. This page provides the details about the evidence that was used to calculate the Total Score for a protein-protein pair. The prediction of protein-protein interaction is calculated using a naive Bayesian classifier. The features that are used to calculate the likelihood of interaction include gene co-expression, orthology to proteins that are known to interact, a combined feature set and a topology module. The combined feature set includes information about co-occurrence of domains, post-translational modifications (PTMs) and co-localisation. The page is split into three columns. The first column is the feature, the second column is the evidence provided by that feature set. The third column shows the score based on the evidence provided. For the co-occurrence of domains, post translational modifications and co-localisation there is only a single score for the three features as these are all part of the combined feature set. FeaturesGene Co-expression: Shows the Pearson's correlation of co-expression over a range of conditions for a protein pair. Orthology: Lists orthologous interactions for a protein pair. An orthologous interaction is when two proteins in one species are inferred to interact due to a homologous pair of proteins in a second species being identified as interacting. Domains: Provides a list of the domains that are present in each of the proteins. Chi square scores for the co-occurrence of the domains are also provided, are also highlighted if the domains are present in one or the other protein or if present in both proteins. Post-Translational Modifications: Lists the PTMs present in both proteins and the residue number of where they occur. Localisation: Lists where the proteins have been annotated to be localised to in the cell. Local Network Topology (Transitive Score): Uses the network topology calculated from gene co-expression, orthology and the combined feature set to predict interaction. The score for interaction is based on a protein pair having a common set of interactions with other proteins. In the table it lists the common interactors between the protein pair and the scores between the protein and the common interactor. The scores listed are those output by the transitive module and not the Total Score value. ExampleThe figure (left) shows the evidence for the prediction of interaction between SNRPG and SNRPD3. The Expression section reports that there is a high Pearson’s correlation of 0.93 between the expression of the two genes as studied in the GDS596 dataset from the NCBI GEO database. The Orthology section shows the orthologous proteins from S. cerevisiae, C. elegans and D. melanogaster as judged by the Inparalog score and highlights the fact that interactions have been documented only in S. cerevisiae, whilst in C. elegans and D. melanogaster the genes have been observed, but no studies have been performed to confirm an interaction. Links to PubMed are included to allow further exploration of the evidence on which the prediction is based. The information required by the combined module covers three sections. The Domains section, lists the domains that are present in each protein along with a Chi-square score for the co-occurrence of the two domains within the protein pair. For post-translational modifications is a list of modifications and the source of the information is shown. The Localisation section shows that both SNRPG and SNRPD3 have been annotated by the HPRD as localising within the nucleus of the cell. The Transitive module makes predictions based on the local predicted network topology by using the interaction score calculated based on only the Expression, Orthology and Combined modules. The Transitive module considers all the common interactors for SNRPG and SNRPD3 that have an score interaction ≥0.025, of which there are 236 in total. The table gives a list of the predicted interactors that have a current interaction score ≥0.025, along with their names and the interaction score between SNRPG and the common interactor and SNRPD3 and the common interactor. For example, both SNRPG and SNRPD3 have a calculated interaction scores of 2.07E3 and 3.41E3, respectively, with the common interactor SNRPD1. |
 |
Protein Summary
This page is linked to by clicking on a protein name from the predicted interactions results table, if an exact sequence match is found in a protein sequence search or from the protein name from the BLAST results page. The table lists the protein name and extended descriptive name, external database reference tags (IPI, RefSeq, HPRD, UniProt and Entrez) and the protein sequence. It also has a break down of the number of predicted interactions in relation to different interaction score cutoff points, the values of which link through to the Protein Interactions results table with the respective cutoff point. |
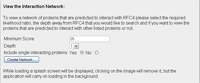  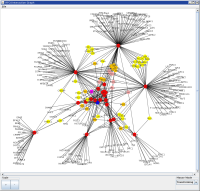 |
Protein Network Viewer
To aid users in being able to visualise the network of predicted protein-protein interactions, there is a Java Web Start application that allows you to view the identified protein and its interacting neighbours. From the Protein Summary page, there is a section to "View the Interaction Network". Here you can:
If the network depth is set to 1, then it will bring back only the interacting proteins of the current protein. If the depth is set to 2 then it will bring back all of the proteins that interact with the current protein provided the interactions have a score greater than the set minimum. When viewing a protein interaction network with a depth greater than 1, selecting "No" will allow you to view only interacting proteins that have more than 1 interaction. Once you have decided on the parameters for the network, click "Create Network ...". This will launch the web application. You will need to click "Run" when you are presented with the security warning. When the application has loaded you can view the network, but also perform the following manipulations on the diagram:
The application requires Java 1.5 or greater to run and uses Java WebStart to load. If you are having difficulties please view the respective help pages at Sun Microsystems for further information about Java: |

