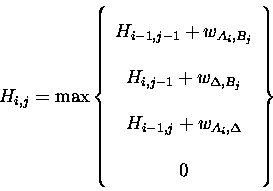 |
(4) |
The general algorithm described in Section 4.3 is a global alignment algorithm. If it is not known in advance that the sequences are similar over most of their length, then a local alignment algorithm is preferred. To convert the basic algorithm to a local algorithm for a pair-score matrix that is centred on zero, the 0th row and column of H are initialised to zero, then the calculation of each element of H is modified to:
The only difference is the addition of a zero [Smith & Waterman, 1981] which has the effect of terminating any path in the H matrix whose score drops below zero. The maximum local alignment score for comparison of Aand B is given by the maximum value in H. The alignment is traced back from this cell until cells on the path drop to zero. There may be more than one local alignment between two sequences, perhaps representing repeats, or multiple shared domains. Algorithms exist to find most [Barton, 1993c] or all [Waterman & Eggert, 1987] of the alternative local alignments.