For Developers¶
Source Code¶
Note
This is an open source project. If you want to contribute or report an issue have a look at our Git-tracker.
Publicly available Git repository: http://source.jalview.org/gitweb/?p=jabaws.git
git clone http://source.jalview.org/git/jabaws.git
The API¶
Data Model JavaDoc - read this if your are coding against JABA Web Services
Complete JavaDoc - for developers who want to use JABAWS framework and use Engines and Executables directly
Structure of the project¶
The code structure¶
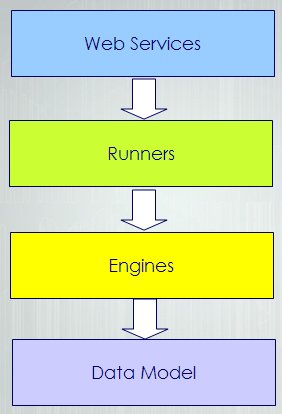
Each source folder depends on the upper folders for compilation. For example, the datamodel is the top level folder so it has no other dependencies on other JABAWS code. The Engine level depends on the datamodel to compile etc. The web services folder is the bottom layer and depends on all the other source code.
So the JABAWS project is split into 4 layers. From bottom-up the first layer consists from the value classes used by all other layers of the hierarchy, in particular web services. So, to be able to use JABAWS one needs to have these classes. At the same time classes on this layer does not have any dependencies on the layers above.
The second layer contains code for execution of the wrappers, which are the abstraction describing native executables. JABAWS can execute tasks locally that is on the same machine as JVM and on the cluster. Thus currently code on this layer contain two engines. This layer depends on the layer underneath, the data model layer, but is completely independent from the code above.
The third layer consists of the wrappers for the native executables and classes to handle their configuration. It depends on the engines and the data model, but know nothing about the web services.
Finally, the upper layer contains the web services, that depend on all the layers below.
The layer isolation is archived though specially designed compilation task which is executed sequentially in several stages so that the first layer compiles before any other layers, second layer compiles after that and process continies before all the code is compiled. Any violation of the layer boundaries results in the compilation failure. Use Ant “Compile” or “Complile_with_debug” tasks to perform the staged compilation.
A client package contains only classes from data model layer and a simple web services client. Framework package is for anyone who want to use JABAWS framework for controlling native executables in local or cluster environments. Framework exclude the web services layer. Server package contains all the code.
Unit Testing¶
JABAWS uses TestNG framework for testing. The test results for the JABAWS package offered for download can be found at: Test Results JABAWS uses TestNG for testing. There is a TestNG plugin available for Eclipse which has functionality similar to JUnit. However, no plugins are necessary to run the test cases, as testng jar is supplied with JABAWS together with an ant tasks to run the test cases.
Several testing groups are supported:
- All tests (‘Test’)
- Cluster tests (‘Run_cluster_dependent_test’)
- Cluster independent tests (‘All_cluster_independent_tests’)
- Windows only tests (‘All_cluster_independent_windows_only_tests’)
- Performance and stability tests (‘Long_tests’)
- Re-run failed tests (‘Rerun_failed_tests’)
- Run custom test (‘CustomTest’)
To run the tests you need to download all sources from repository. Once you have done that, enter into the command line mode, change directory to the project directory and type:
ant -f build.xml <test group name>
Make sure you have Apache Ant installed and path to ant executable is defined in your path environmental variable. Replace test group name with the one of the names given in the list above to run required group of tests e.g for running cluster only tests use the following command:
ant -f build.xml Run_cluster_dependent_test
If you work under Linux you could use a simple script from the root folder of repository called runtests.sh. This script simply contains a collection of the test commands described above and paths to java home directory and an ant executable, which you can define once for your system and then reuse.
A handy feature of TestNG is its ability to re-run failed tests. Failed test ant file is stored in test-output/testng-failed.xml. and is used in the ant task called Rerun_failed_tests. So re-running failed tests requires no more work than running any other test group and could be accomplished with the command:
ant -f build.xml Rerun_failed_tests
CustomTest runs the test defined in the project root directory file called temp-testng-customsuite.xml. This file is generated by TestNG plugin every time you run the test from Eclipse. Thus an easy way to run a test in a different environment is to run it from Eclipse first and then from ant using a custom test procedure.
For cluster execution make sure that the property LD_LIBRARY_PATH defined in build.xml points to cluster engine LD libraries directory in your local system.
Accessing JABAWS from your program¶
Web services functions overview¶
All JABAWS multiple sequence alignment web services comply to the same interface, thus the function described below are available from all the services.
Functions for initiating the alignment
String id = align(List<FastaSequence> list)
String id = customAlign(List<FastaSequence> sequenceList, List<Option> optionList)
String id = presetAlign(List<FastaSequence> sequenceList, Preset preset)
Functions pertaining to job monitoring and control
JobStatus status = getJobStatus(String id)
Alignment al = getResult(String id)
boolean cancelled = cancelJob(String id)
ChunkHolder chunk = pullExecStatistics(String id, long marker)
Functions relating to service features discovery
RunnerConfig rc = getRunnerOptions()
Limit limit = getLimit(String name)
LimitsManager lm = getLimits()
PresetManager pm = getPresets()
Please refer to a Data Model JavaDoc for a detailed description of each methods.
Structure of the template command line client¶
| Packages | Classes and Interfaces |
|---|---|
| compbio.data.msa | MsaWS the interface for all multiple sequence alignment web services |
| compbio.data.sequence | JABAWS data types |
| compbio.metadata | JABAWS meta data types |
| compbio.ws.client | JABAWS command line client |
Additional utility libraries that this client depend upon is the compbio-util-1.3.jar and compbio-annotation-1.0.jar.
Please refer to a Data Model JavaDoc for a detailed description of each class and its methods.
Connecting to JABAWS¶
For a complete working example of JABAWS command line client please see compbio.ws.client.Jws2Client class. JABAWS command line client source code is available from the download page. Please note that for now all the examples are in Java, other languages will follow if there is sufficient demand.
Download a binary JABAWS client. Add the client to the class path. The following code excerpt will connect your program to Clustal web service deployed in the University of Dundee.
import java.net.URL;
import javax.xml.namespace.QName;
import javax.xml.ws.Service;
// (...)
String qualifiedName = "http://msa.data.compbio/01/01/2010/";
URL url = new URL("http://www.compbio.dundee.ac.uk/jabaws/ClustalWS?wsdl");
QName qname = new QName(, "ClustalWS");
Service serv = Service.create(url, qname);
MsaWS msaws = serv.getPort(new QName(qualifiedName, "ClustalWSPort"),
MsaWS.class);
Line 1 makes a qualified name for JABA web services.
Line 2 constructs the URL to the web services WSDL.
Line 3 makes a qualified name instance for Clustal JABA web service.
Line 4 creates a service instance.
Line 5 makes a connection to the server.
A more generic connection method would look like this
import java.net.URL;
import javax.xml.namespace.QName;
import javax.xml.ws.Service;
import compbio.ws.client.Services
// (...)
String qualifiedServiceName = "http://msa.data.compbio/01/01/2010/";
String host = "http://www.compbio.dundee.ac.uk/jabaws";
// In real life the service name can come from args
Services clustal = Services.ClustalWS;
URL url = new URL(host + "/" + clustal.toString() + "?wsdl");
QName qname = new QName(qualifiedServiceName, clustal.toString());
Service serv = Service.create(url, qname);
MsaWS msaws = serv.getPort(new QName(qualifiedServiceName, clustal + "Port"), MsaWS.class);
Where Services is enumeration of JABAWS web services. All JABAWS multiple sequence alignment methods confirm to MsaWS specification, thus from the caller point of view all JABAWS web services can be represented by MsaWS interface. The full documentation of MsaWS functions is available from the JavaDoc.
Aligning Sequences¶
Given that msaws is web service proxy, created as described in “Connecting to JABAWS” section, the actual alignment can be obtained as follows:
List<FastaSequence> fastalist = SequenceUtil.readFasta(new FileInputStream(file));
String jobId = msaws.align(fastalist);
Alignment alignment = msaws.getResult(jobId);
Line one loads FASTA sequence from the file.
Line two submits them to web service represented by msaws proxy.
Line three retrieves the alignment from a web service. This line will block the execution until the result is available. Use this with caution. In general, you should make sure that the calculation has been completed before attempting retrieving results. This is to avoid keeping the connection to the server on hold for a prolonged periods of time. While this may be ok with your local server, our public server (www.compbio.dundee.ac.uk/jabaws) will not let you hold the connection for longer than 10 minutes. This is done to prevent excessive load on the server. The next section describes how to check the status of the calculation. Methods and classes mentioned in the excerpt are available from the JABAWS client library.
Checking the status of the calculation¶
You may have noticed that there was no pause between submitting the job and retrieving of the results. This is because getResult(jobId) method block the processing until the calculation is completed. However, taking into account that the connection holds server resources, our public server (www.compbio.dundee.ac.uk/jabaws) is configured to reset the connection after 10 minutes of waiting. To work around the connection reset you are encouraged to check whether the calculation has been completed before accessing the results. You can do it like this:
while (msaws.getJobStatus(jobId) != JobStatus.FINISHED) {
Thread.sleep(2000); // wait two seconds, then recheck the status
}
Aligning with presets¶
PresetManager presetman = msaws.getPresets();
Preset preset = presetman.getPresetByName(presetName);
List<FastaSequence> fastalist = SequenceUtil.readFasta(new FileInputStream(file));
String jobId = msaws.presetAlign(fastalist, preset);
Alignment alignment = msaws.getResult(jobId);
Line one obtains the lists of presets supported by a web service.
Line two return a particular Preset by its name.
Lines three to five are doing the same job as in the first aligning sequences example.
Aligning with custom parameters¶
RunnerConfig options = msaws.getRunnerOptions();
Argument matrix = options.getArgument("MATRIX");
matrix.setValue("PAM300");
Argument gapopenpenalty = options.getArgument("GAPOPEN");
gapopenpenalty.setValue("20");
List<Argument> arguments = new ArrayList<Argument>();
arguments.add(matrix); arguments.add(gapopenpenalty);
List<FastaSequence> fastalist = SequenceUtil.readFasta(new FileInputStream(file));
String jobId = msaws.customAlign(fastalist, arguments);
Alignment alignment = msaws.getResult(jobId);
Line one obtains the RunnerConfig object that holds information on supported parameters and their values
Line two retrieve a particular parameter from the holder by its name.
Lines three sets a value to this parameter which will be used in the calculation.
Line four and five do the same but for another parameter.
Line six makes a List to hold the parameters.
Line seven puts the parameters into that list.
Line eight and ten is the same as in previous examples.
Line nine submit an alignment request with the sequences and the parameters.
The names of all the parameters supported by a web service e.g. “PAM300” can be obtained using options.getArguments() method. Further details on the methods available from RunnerConfig object are available from the JavaDoc.
Writing alignments to a file¶
There is a utility method in the client library that does exactly that.
Alignment alignment = align(...)
FileOutputStream outStream = new FileOutputStream(file);
ClustalAlignmentUtil.writeClustalAlignment(outStream, align);
A complete client example¶
Finally, a complete example of the program that connects to JABAWS Clustal service and aligns sequences using one of the Clustal web service presets. All you need for this to work is a JABAWS CLI client. Please make sure that the client is in the Java class path before running this example.
import java.io.ByteArrayInputStream;
import java.io.FileNotFoundException;
import java.io.IOException;
import java.net.URL;
import java.util.List;
import javax.xml.namespace.QName;
import javax.xml.ws.Service;
import compbio.data.msa.MsaWS;
import compbio.data.sequence.Alignment;
import compbio.data.sequence.FastaSequence;
import compbio.data.sequence.SequenceUtil;
import compbio.metadata.JobSubmissionException;
import compbio.metadata.LimitExceededException;
import compbio.metadata.Preset;
import compbio.metadata.PresetManager;
import compbio.metadata.ResultNotAvailableException;
import compbio.metadata.UnsupportedRuntimeException;
import compbio.metadata.WrongParameterException;
public class Example {
/*
* Input sequences for alignment
*/
static final String input = ">Foo\r\n"
+ "MTADGPRELLQLRAAVRHRPQDFVAWLMLADAELGMGDTTAGEMAVQRGLALHPGHPEAVARLGR"
+ "VRWTQQRHAEAAVLLQQASDAAPEHPGIALWLGHALEDAGQAEAAAAAYTRAHQLLPEEPYITAQ"
+ "LLNWRRRLCDWRALDVLSAQVRAAVAQGVGAVEPFAFLSEDASAAEQLACARTRAQAIAASVRPL"
+ "APTRVRSKGPLRVGFVSNGFGAHPTGLLTVALFEALQRRQPDLQMHLFATSGDDGSTLRTRLAQA"
+ "STLHDVTALGHLATAKHIRHHGIDLLFDLRGWGGGGRPEVFALRPAPVQVNWLAYPGTSGAPWMD"
+ "YVLGDAFALPPALEPFYSEHVLRLQGAFQPSDTSRVVAEPPSRTQCGLPEQGVVLCCFNNSYKLN"
+ "PQSMARMLAVLREVPDSVLWLLSGPGEADARLRAFAHAQGVDAQRLVFMPKLPHPQYLARYRHAD"
+ "LFLDTHPYNAHTTASDALWTGCPVLTTPGETFAARVAGSLNHHLGLDEMNVADDAAFVAKAVALAS"
+ "DPAALTALHARVDVLRRESGVFEMDGFADDFGALLQALARRHGWLGI\r\n"
+ "\r\n"
+ ">Bar\r\n"
+ "MGDTTAGEMAVQRGLALHQQRHAEAAVLLQQASDAAPEHPGIALWLHALEDAGQAEAAAAYTRAH"
+ "QLLPEEPYITAQLLNAVAQGVGAVEPFAFLSEDASAAESVRPLAPTRVRSKGPLRVGFVSNGFGA"
+ "HPTGLLTVALFEALQRRQPDLQMHLFATSGDDGSTLRTRLAQASTLHDVTALGHLATAKHIRHHG"
+ "IDLLFDLRGWGGGGRPEVFALRPAPVQVNWLAYPGTSGAPWMDYVLGDAFALPPALEPFYSEHVL"
+ "RLQGAFQPSDTSRVVAEPPSRTQCGLPEQGVVLCCFNNSYKLNPQSMARMLAVLREVPDSVLWLL"
+ "SGPGEADARLRAFAHAQGVDAQRLVFMPKLPHPQYLARYRHADLFLDTHPYNAHTTASDALWTGC"
+ "PVLTTPGETFAARVAGSLNHHLGLDEMNVADDAAFVAKAVALASDPAALTALHARVDVLRRESGV"
+ "FEMDGFADDFGALLQALARRHGWLGI\r\n"
+ "\r\n"
+ ">Friends\r\n"
+ "MTADGPRELLQLRAAVRHRPQDVAWLMLADAELGMGDTTAGEMAVQRGLALHPGHPEAVARLGRV"
+ "RWTQQRHAEAAVLLQQASDAAPEHPGIALWLGHALEDHQLLPEEPYITAQLDVLSAQVRAAVAQG"
+ "VGAVEPFAFLSEDASAAEQLACARTRAQAIAASVRPLAPTRVRSKGPLRVGFVSNGFGAHPTGLL"
+ "TVALFEALQRRQPDLQMHLFATSGDDGSTLRTRLAQASTLHDVTALGHLATAKHIRHHGIDLLFD"
+ "LRGWGGGGRPEVFALRPAPVQVNWLAYPGTSGAPWMDYVLGDAFALPPALEPFYSEHVLRLQGAF"
+ "QPSDTSRVVAEPPSRTQCGLPEQGVVLCCFNNSYKLNPQSMARMLAVLREVPDSVLWLLSGPGEA"
+ "DARLRAFAHAQGVDAQRLVFMPKLPHPQYLARYRHADLFLDTHPYNAHTTASDALWTGCPVLTTP"
+ "GETFAARVAGSLNHHLGLDEMNVADDAAFVAKAVALASDPAALTALHARVDVLRRESI";
public static void main(String[] args) throws UnsupportedRuntimeException,
LimitExceededException, JobSubmissionException,
WrongParameterException, FileNotFoundException, IOException,
ResultNotAvailableException, InterruptedException {
String qualifiedServiceName = "http://msa.data.compbio/01/01/2010/";
/* Make a URL pointing to web service WSDL */
URL url = new URL("http://www.compbio.dundee.ac.uk/jabaws/ClustalWS?wsdl");
/*
* If you are making a client that connects to different web services
* you can use something like this:
*/
// URL url = new URL(host + "/" + Services.ClustalWS.toString() +
// "?wsdl");
QName qname = new QName(qualifiedServiceName, "ClustalWS");
Service serv = Service.create(url, qname);
/*
* Multiple sequence alignment interface for Clustal web service
* instance
*/
MsaWS msaws = serv.getPort(new QName(qualifiedServiceName, "ClustalWS"
+ "Port"), MsaWS.class);
/* Get the list of available presets */
PresetManager presetman = msaws.getPresets();
/* Get the Preset object by preset name */
Preset preset = presetman
.getPresetByName("Disable gap weighting (Speed-oriented)");
/*
* Load sequences in FASTA format from the file You can use something
* like new FileInputStream(<filename>) to load sequence from the file
*/
List<FastaSequence> fastalist = SequenceUtil
.readFasta(new ByteArrayInputStream(input.getBytes()));
/*
* Submit loaded sequences for an alignment using preset. The job
* identifier is returned by this method, you can retrieve the results
* with it sometime later.
*/
String jobId = msaws.presetAlign(fastalist, preset);
/* This method will block for the duration of the calculation */
Alignment alignment = msaws.getResult(jobId);
/*
* This is a better way of obtaining results, it does not involve
* holding the connection open for the duration of the calculation,
* Besides, as the University of Dundee public server will reset the
* connection after 10 minutes of idling, this is the only way to obtain
* the results of long running task from our public server.
*/
// while (msaws.getJobStatus(jobId) != JobStatus.FINISHED) {
// Thread.sleep(1000); // wait a second, then recheck the status
// }
/* Output the alignment to standard out */
System.out.println(alignment);
// Alternatively, you can record retrieved alignment into the file in
// ClustalW format
// ClustalAlignmentUtil.writeClustalAlignment(new FileOutputStream(
// "output.al"), alignment);
}
}
For a more detailed description of all available types and their functions please refer to the Data Model JavaDoc.
Adding new web-services¶
Brief Guide¶
Add a new executable which you’d like to wrap as a JABAWS web service to the binaries folder. If it has the source code and can be recompiled for different platforms include it under
binaries/src. Editsetexecutableflag.shandcompilebin.shscripts inbinaries/srcaccordingly.Make sure that all the dependencies of the software being installed are satisfied. If there are other binaries they should be included as well. Keep the dependent binaries in a subfolder for the main executable. Update
compilebin.shandsetexecflag.shscripts accordingly.Make sure that the new executable does not have any hard links to its dependencies, e.g. is able to run from any installation folder and does not contain any hard coded paths.
Describe executable in
conf/Exectuable.propertiesfile. The lowercase name of the wrapper should be included in the name of the property for example Clustal properties all include clustal as a part of the name e.g.local.clustalw.bin. The same property for MAFFT will be calledlocal.mafft.bin. For more help please refer to the Executable.properties file.Describe the executable supported parameters in the
<ExecutableName>Parameters.xml, presets in the<ExecutableName>Presets.xmland the execution limits in the<ExecutableName>Limit.xml. By convention these files are stored inconf/settings. All of these are optional. If the executable does not support parameters you do not have to mention theXXXParameter.xmlfile in theExecutable.propertiesfile at all. The same is true for Presets and Limits.Create a Java wrapper class for your executable. Create it within runner source directory. Examples of other wrappers can be found in
compbio.runner.msaor in othercompbio.runner.*packages. Wrapper should extendSkeletalExecutable<T>and implementPipedExecutable<T>if you need to pass the input or collect the results from the standard in/out. Please see Mafft code as example. Wrapper should expendSkeletalExecutable<T>if input/output can be set as a parameter for an executable. Please see the ClustalW code as example.Create a testcase suit for your wrapper in
testsrcand run the test cases.Create parser for the output files of your executable. Suggested location
compbio.data.sequence.SequenceUtil.Test the parser.
Decide which web services interfaces your executable is going to match. For example if the executable output can be represented as SequenceAnnotation then SequenceAnnotation interface might be appropriate. For multiple sequence alignment an Msa interface should be used.
If you find a web interface that matches your returning data type, then implement a web service which confirms to it within a webservices source folder.
Register web service in
WEB-INF/web.xmlandWEB-INF/sun-jaxws.xml.Add generated wsdl to wsbuild.xml ant script to generate the stubs.
Run build-server task in wsbuild file. Watch for errors. If the task fails that means that JAXB cannot serialize some of your new data structures. Add appropriate annotations to your data types. Also check that:
- you do not have interfaces to serialize, since JAXB cannot serialize them
- you have a default no args constructor (can be private if you do not need it)
- JAXB cannot serialize Java Map class, use a custom data structure instead
- Enum cannot be serialized as its abstract class (do not confuse with enum which is fine)
- Fields serialization leaves a little more space for manoeuvre. If you do this then you may accept and return interfaces, e.g. List, Map; abstract classes etc, from your methods
If you have the data on the server side, but nothing is coming through to the client, this is a JAXB serialization problem. They tend to be very silent and thus hard to debug. Check your data structure can be serialized!
Modify the client to work with your new web service. Update Services enumeration to include new service and ensure that all the methods of this enumeration take into account the new service. Update the client help text (
client_help.txt) and insert it into the Constraints class.Test the web service with the client.
Test on the cluster.
Building web services artifacts¶
JABAWS are the standard JAX-WS SOAP web services, which are WS-I basic profile compatible. This means that you could use whatever tool your language has to work with web services. Below is how you can generate portable artifacts to work with JABAWS from Java. However if programming in Java, we recommend using our client library as it provides a handful of useful methods in addition to plain data types.
wsimport -keep http://www.compbio.dundee.ac.uk/jabaws/ClustalWS?wsdl
Server side artifacts should be rebuild whenever the data model, meta model or MSA interface were changed. To do that run build-server task from wsbuild.xml ant build file. WSDL files will be generated in webservices/compbio/ws/server/resource directory. It is not necessary to edit them if any of the JABAWS clients are used. JABAWS are the standard JAX-WS web services, which are WS-I basic profile compatible.
Preparing Distributives¶
There are a number of ant tasks aimed for preparing distributives for download. Currently a few types of JABAWS packages are offered:
- Client only (contains classes required to access JABA Web Services)
- Platform specific JABAWS (windows and other)
- JABAWS with and without binaries
- JABAWS framework and complete project
The easiest way to build all distributives is to call build-all ant task. There are more tasks defined in build.xml than described here. They are mostly self explanatory.
If you made any changes to the data model and would like to generate a complete JABAWS distro make sure you have rebuilt jaxws artifact as described below.
